Where Evolution meets Connectomics, Transcriptomics, and Tool Development.
How do you build a brain? How does a primordial brain circuit adapt to changing functions? And what do answers to such evolutionary questions teach us about the organizing principles of the vertebrate brain and its function in health and disease?
To understand how brain regions, cell types, and their connections evolved and function, we are taking a comparative approach to connectomics and transcriptomics. We develop and apply molecular tools — including MAPseq, cellular barcoding, in situ sequencing, single-cell RNA and ATACseq, viral genetic tracing, brain clearing, and light-sheet imaging — to map the brains of mice and other mammals, birds, reptiles, and amphibians faster and at higher resolution than ever before. Comparing and contrasting these datasets then reveals conserved and specialized circuit features and ultimately allows us to derive principles of vertebrate brain organization. Developmental investigations supply the mechanisms for the observed circuit differences.
In parallel to these evolutionary studies, we apply our tools to models of developmental disorders, neurodegeneration, and addiction to test how brain structure changes over disease progression and how the organizing principles of the vertebrate brain are violated in disease.
We are always looking for Postdocs, PhD Students, and technicians to join us! Find out more below.
Brain Evolution
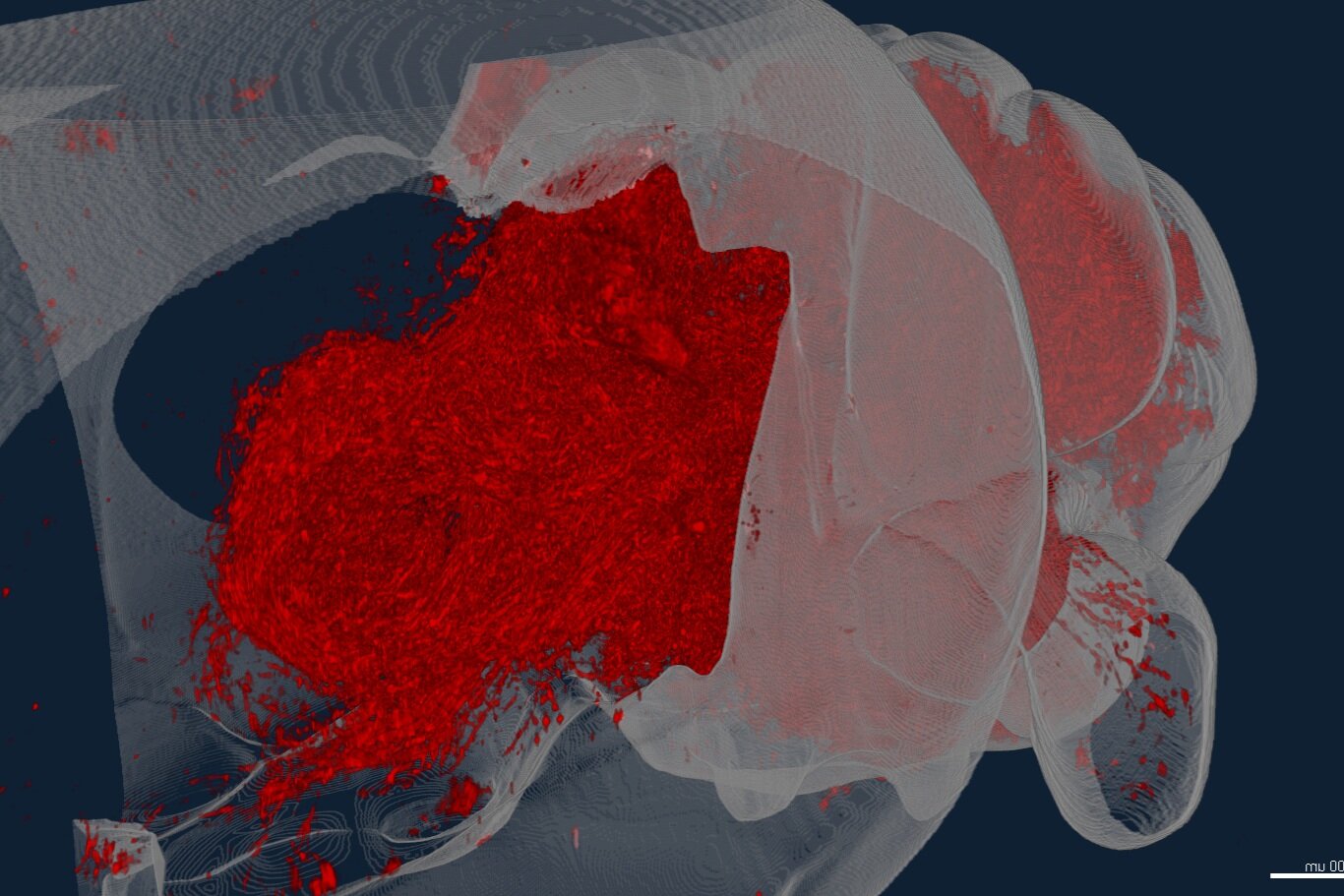
We are studying how circuits change and expand across evolution at the resolution of cell types, brain regions and brain-wide connectivity. In the cerbellum, we recently discovered a deeply conserved set of cell types that form an archetypal cerebellar nucleus, and suggest that new brain areas form by a process of duplication-and-divergence. In the lab, we are testing how these processes are implemented in development and how they generalize to other brain areas.
Barcoding the Brain
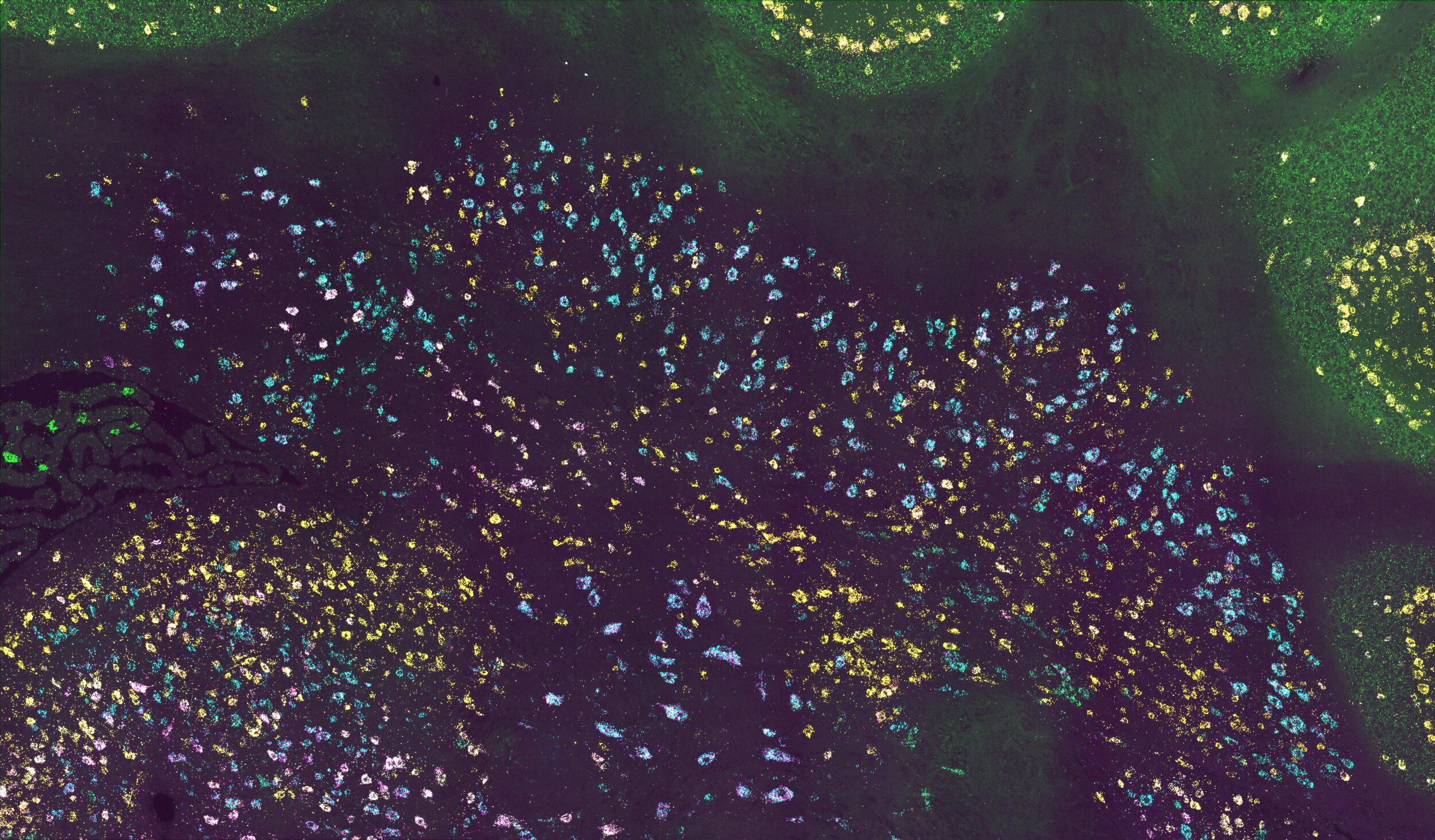
To map brains quickly and at single-cell resolution, we developed tools like MAPseq and BRICseq that translate the problem of neuroanatomy into a problem solvable by high-throughput sequencing. Using these tools we can map tens of thousands of individual neurons, or entire mesoscale connectomes in individual animals. We now develop the next generation of barcoding tools and leverage this technology to bring high-resolution connectomics to non-standard model species, paving the way for high-throughput comparative connectomics.
Viral Engineering
Viruses are indispensable tools in circuit mapping. Nature evolved different viruses to move through the nervous system in different ways. We exploit these natural characteristics and combine them with molecular trickery and cellular barcoding to maximize the information return of any experiment, and teach us new things about brain connectivity.
Selected Publications
Kim H, Xu C, Washington C, Lowman M, Kebschull JM. Cell type-specific barcoding reveals the projectional architecture of the mouse midbrain dopaminergic system. bioRxiv 2025.06.25.661405. PDF
Kim H, Qi H, Washington C, Liang X, Kebschull JM. MAPseq2: a sensitive and cost-effective barcoded connectomics method. bioRxiv 2025.06.23.661165. PDF
Banerjee A, Phelps SM, Kebschull JM. The Unusual Effectiveness of Evolution in Systems Neuroscience. Cold Spring Harb Perspect Biol. 2025 Sep 22:a041510. doi: 10.1101/cshperspect.a041510. PDF
Kebschull JM, Casoni F, Consalez GG, Goldowitz D, Hawkes R, Ruigrok TJH, Schilling K, Wingate R, Wu J, Yeung J, Uusisaari MY. Cerebellum Lecture: the Cerebellar Nuclei-Core of the Cerebellum. Cerebellum. 2023 Feb 13. PDF
Kebschull JM, Richman EB, Ringach N, Friedmann D, Albarran E, Kolluru SS, Jones RC, Allen WE, Wang Y, Cho SW, Zhou H, Ding JB, Chang HY, Deisseroth K, Quake SR, Luo L. Cerebellar nuclei evolved by repeatedly duplicating a conserved cell-type set. Science 370, eabd5059 (2020). DOI: 10.1126/science.abd5059 PDF
Huang L*, Kebschull JM*, Furth D, Musall S, Kaufman MT, Churchland AK, Zador AM. BRICseq bridges brain-wide interregional connectivity to neural activity and gene expression in single animals. Cell. 2020; 182(1), 117-188.e7. PDF
Han Y*, Kebschull JM*, Campbell RAA*, Cowan D, Imhof F, Zador AM, Mrsic-Flogel TM. The logic of single-cell projections from visual cortex. Nature. 2018; 556(7699), 51-56. PDF
Kebschull JM, Garcia da Silva P, Reid AP, Peikon ID, Albeanu DF, Zador AM. High-throughput mapping of single-neuron projections by sequencing of barcoded RNA. Neuron. 2016; 91(5), 975-987. PDF